Adverse Effects of Wheat Gluten
Key insights
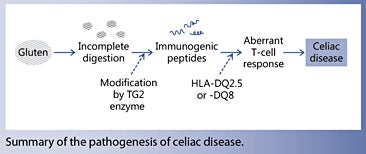
Celiac disease is a chronic autoimmune enteropathy that is influenced by environmental and genetic factors. It is the only gastrointestinal disorder in which the involvement of gluten has been clearly established. Due to its high proline content, gluten is resistant to degradation in the gastrointestinal tract. A key pathogenic event in celiac disease is the aberrant T-cell responses directed against gluten fragments. These antigenic fragments can activate immune cells in the gut, recruit infiltrating T cells, and initiate proinflammatory signalling events that eventually lead to profound remodelling of the gut tissues.
Current knowledge
Wheat gluten consists of two classes of proteins, the gliadins and the glutenins. The glutenins confer elasticity and viscosity to various food products, but the immunogenic peptide fragments that trigger symptoms in patients with celiac disease come from the gliadin family. The majority of patients with celiac disease express either HLA-DQ2.5 and/or HLA-DQ8. Under normal conditions, these molecules bind and present antigen-derived peptides to T cells. In celiac disease, aberrant T-cell responses are directed towards gluten fragments bound to HLA-DQ2.5 and/or HLA-DQ8. Such immunogenic fragments arise from partial degradation of gluten proteins in the gastrointestinal tract, which are then modified by the tissue transglutaminase (TG2) enzyme.
Practical implications
Patients with celiac disease should avoid food products that contain wheat, barley or rye. In general, oat is considered safe for patients with celiac disease. Oral supplements of post-proline digesting enzymes (prolyl endopeptidases and prolyl endoproteases) present one strategy for enhancing gluten degradation. Although this alone will not enable patients to consume a normal gluten-containing diet, use of these enzymes may be effective at counteracting the effects of inadvertent gluten exposure.
Recommended reading
Tjon JM, van Bergen J, Koning F: Celiac disease: how complicated can it get? Immunogenetics 2010;62:641–651.
- Celiac disease (and the associated dermatitis herpetiformis) is the only (gastrointestinal) disorder where the involvement of gluten has been clearly established.
- The unique properties of gluten proteins explain their disease-inducing properties.
- Safe wheat and safe gluten are not yet within reach, but the current knowledge allows alternative approaches to meet patients’ needs.
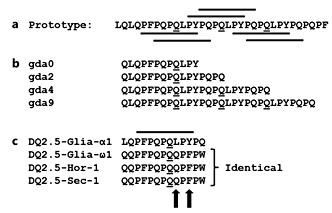
The special amino acid composition of gluten is strongly linked to its disease-inducing capacity. All gluten proteins are glutamine (symbol Q) and proline (symbol P) rich. Together, these two amino acids make up almost 50% of the gluten proteins. Due to their proline-rich nature, the gluten proteins are difficult to degrade, hence they persist in the gastrointestinal tract [4]. Also, many HLA-DQ2.5-restricted gluten epitopes have a proline at relative position 8 which is crucial for T-cell recognition [17, 18]. However, perhaps most importantly, its prolineand glutamine-rich nature makes gluten a perfect substrate for the enzyme tissue transglutaminase [5], which generates gluten fragments that bind with high affinity to either HLA-DQ2.5 or HLA-DQ8, a prerequisite for T-cell recognition of most immunogenic gluten fragments.
Both HLA-DQ2.5 and HLA-DQ8 preferentially bind peptides with a negatively charged amino acid at relative position 4, 6 or 7 (HLA-DQ2.5) or at position 1 and/or 9 (HLA-DQ8), but gluten does not contain negatively charged amino acids. This discrepancy was solved when it was realized that the immunogenic gluten peptides contained one or more glutamine residues at critical positions and that replacing these amino acids by glutamic acid, which is negatively charged, allowed for high-affinity binding of the modified peptides to either HLADQ2.5 or -DQ8. Importantly, it was shown that such a modification of gluten peptides is mediated by the enzyme tissue transglutaminase (TG2), which under normal circumstances is located intracellularly where it is enzymatically inactive but becomes active when it is released upon tissue damage [7, 16]. Moreover, QLPY and QLPF sequences, which are abundantly present in gluten proteins, were found to be an ideal substrate for TG2 and are converted in ELPY and ELPF, where the E stands for the negatively charged glutamic acid [5]. Consequently, in the 33-mer, three Q residues are converted into an E, yielding 6 peptides that bind with high affinity to HLADQ2.5 due to the introduction of a negative charge at either position 4 or 6 (fig. 3 , 4). Similarly, TG2 can introduce negative charges at position 1 and/or 9 in other gluten peptides, which generates peptides that bind with high affinity to HLA-DQ8, like in the case of the DQ8- glia-α1 peptide [16] (table 1). Thus, the unique amino acid composition of gluten explains its immunogenic nature.
While it is clear that most gluten toxicity is associated with the α- and ω-gliadins, T-cell epitopes have also been identified in the γ-gliadins and in the LMW and HMW glutenins. In fact, T-cell responses to the γ-gliadins are quite frequently detected in patients, while the glutenins appear to have a much lower toxicity profile. Notably, the identified γ-gliadin and glutenin epitopes are less proline rich (2–3 per epitope) compared to the epitopes in the α- and ω-gliadins (4 per epitope). This may render the γ-gliadin and glutenin epitopes more susceptible to enzymatic degradation which could relate to their lower immunogenicity.
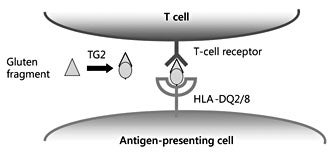
Recent studies have revealed the molecular basis for the interaction between public T-cell receptors and HLADQ gluten [21–23]. In both HLA-DQ2 and HLA-DQ8 disease, it was observed that this interaction is mediated by a number of key interactions between the T-cell receptors, HLA-DQ and the bound gluten peptide. Strikingly, in all cases, an arginine residue from the T-cell receptor was found to play a crucial role. Thus, high-affinity recognition of HLA-DQ gluten is mediated by a relatively conserved set of T-cell receptors that predominate in patients and appear absent in healthy individuals. Elimination of such T cells might thus constitute an effective therapeutic option for patients with CD.
These antibodies are directed to a repetitive sequence in gluten: QPFXXQXPY, where X can be any amino acid [29]. Such a sequence is also present in the p31-43 peptide: 31-LGQQQPFPPQQPY-43, implying that the 33- peptide could be targeted by gluten-specific antibodies. As the p31-43 peptide is directly adjacent to the immunogenic 33-mer, uptake of large gliadin fragments containing both p31-43 and the 33-mer might thus be facilitated by such antibodies expressed on gliadin-specific B cells or by antigen-presenting cells expressing Fc receptors. In both cases, this would result in intracellular processing of the internalized gliadin fragment, which would allow binding to HLA-DQ and transport of the HLA-DQgluten complexes to the cell surface. Thus, the antibodymediated gliadin uptake would lead to strongly enhanced HLA-DQ-mediated presentation of gliadin to gliadin- specific T cells and promote inflammation. Conversely, the T cells would provide ‘help’ to the antibodyproducing B cells, leading to enhanced antibody production and creating a powerful amplification loop. Thus, the immune modulatory effects of the p31-43 peptide might be indirect and explained by enhanced adaptive T-cell responses.
On a more positive note, evidence has been provided that natural variants of immunogenic gluten fragments exist that lack T-cell stimulatory properties [17, 18]. In many cases, these variant peptides differ in only one amino acid from their immunogenic counterpart and this often concerns a proline to serine (symbol S) substitution at relative position 8. For example, the DQ2-glia-α2 epitope PQPQLPYPQ from the D-genome is highly immunogenic, while its naturally occurring counterpart PQPQLPYSQ from the A-genome is not [17, 18]. Thus, gliadin genes could be engineered so that they would no longer encode immunogenic sequences while retaining most of their natural properties. However, due to the repetitive nature of the α-gliadins, multiple proline residues would need to be modified to eliminate all toxicity. This would involve six substitutions in the 33-mer alone, complicating such an approach. Therefore, although strategies to produce safe gluten exist, it would be very expensive and thus at present not a realistic option.
The latter enzyme, AN-PEP, has a favorable pH profile as it is highly active at low pH values and it is resistant to degradation by pepsin [32]. Consequently, it is capable of degrading gluten in the stomach compartment both in vitro and in vivo and it has been found to exert no harmful effects in patients and healthy human volunteers [32, 35–37]. The enzyme is expected to reach the market as an over-the-counter product in 2015. In addition, S. capsulate PEP has been combined with a cysteine endoprotease B from barley; the latter cleaves after glutamine [38] . The combined activity of these two enzymes is highly effective in degrading gluten as well, and the efficacy is currently being tested in a phase IIb study.
It is doubtful, however, that the availability of such enzyme preparations will allow patients to eat a normal gluten- containing diet. Daily gluten intake varies between 10 and 20 g in adults on a gluten-containing diet. Gluten is present in many food products, and any intake of gluten would have to be accompanied by oral administration of gluten-degrading enzymes. Moreover, evidence has been provided that the efficacy of gluten degradation depends on the context in which gluten is consumed. In the case of AN-PEP, for example, artificial lowering of the pH by adding a carbonated drink enhanced gluten degradation while milk slowed the process [39]. It is therefore hard to imagine that any enzyme will be effective in ensuring complete degradation of the high amounts of gluten protein present in a normal diet before it reaches the small intestine. The enzymes, however, would be quite effective in degrading relative small amounts of gluten and could thus be used to counteract the deleterious effect of inadvertent gluten exposure, for example when eating out or on holidays.
- Shewry P: Wheat. J Exp Bot 2009; 60: 1537–1553.
- Spaenij-Dekking EHA, Kooy Y, Koning F: The Ethiopian cereal tef in celiac disease. N Engl J Med 2005; 353: 1748–1749.
- Sollid LM, Qiao SW, Anderson RP, Gianfrani C, Koning F: Nomenclature and listing of celiac disease relevant gluten T-cell epitopes restricted by HLA-DQ molecules. Immunogenetics 2012; 64: 455–460.
- Shan L, Molberg O, Parrot I, Hausch F, Filiz F, Gray GM, Sollid LM, Khosla C: Structural basis for gluten intolerance in celiac sprue. Science 2002; 297: 2275–2279.
- Vader W, de Ru A, van der Wal Y, Kooy Y, Benckhuijsen W, Mearin L, Drijfhout JW, van Veelen P, Koning F: Specificity of tissue transglutaminase explains cereal toxicity in celiac disease. J Exp Med 2002; 195: 643–649.
- van de Wal Y, Kooy Y, van Veelen P, Pena S, Mearin L, Molberg O, Lundin K, Sollid L, Mutis T, Benckhuijsen W, Drijfhout JW, Koning F: Small intestinal cells of celiac disease patients recognize a natural pepsin fragment of gliadin. Proc Natl Acad Sci USA 1998; 95: 10050–10054.
- Molberg O, McAdam SN, Korner R, Quarsten H, Kristiansen C, Madsen L, Fugger L, Scott H, Noren O, Roepstorff P, Lundin KE, Sjostrom H, Sollid LM: Tissue transglutaminase selectively modifies gliadin peptides that are recognized by gut-derived T cells in celiac disease. Nat Med 1998; 4: 713–717.
- Anderson RP, Degano P, Godkin AJ, Jewell DP, Hill AV: In vivo antigen challenge in celiac disease identifies a single transglutaminase-modified peptide as the dominant A-gliadin T-cell epitope. Nat Med 2000; 6: 337–342.
- Arentz-Hansen H, Körner R, Molberg O, Quarsten H, Vader W, Kooy YM, Lundin KEA, Koning F, Roepstorff P, Sollid LM, Mc-Adam SN: The intestinal T cell response to α-gliadin in adult celiac disease is focused on a single deamidated glutamine targeted by tissue transglutaminase. J Exp Med 2000; 191: 603–612.
- Vader W, Kooy Y, van Veelen P, de Ru A, Harris D, Benckhuijsen W, Pena S, Mearin L, Drijfhout JW, Koning F: The gluten response in children with recent onset celiac disease. A highly diverse response towards multiple gliadin and glutenin derived peptides. Gastroenterology 2002; 122: 1729–1737.
- Tye-Din JA, Stewart JA, Dromey JA, Beissbarth T, van Heel DA, Tatham A, Henderson K, Mannering SI, Gianfrani C, Jewell DP, Hill AV, McCluskey J, Rossjohn J, Anderson RP: Comprehensive, quantitative mapping of T cell epitopes in gluten in celiac disease. Sci Transl Med 2010; 2: 41ra51.
- Tjon JM, van Bergen J, Koning F: Celiac disease: how complicated can it get? Immunogenetics 2010; 62: 641–651.
- Abadie V, Sollid LM, Barreiro LB, Jabri B: Integration of genetic and immunological insights into a model of celiac disease pathogenesis. Annu Rev Immunol 2011; 29: 493–525.
- Christophersen A, Ráki M, Bergseng E, Lundin KE, Jahnsen J, Sollid LM, Qiao SW: Tetramer-visualized gluten-specific CD4+ T cells in blood as a potential diagnostic marker for coeliac disease without oral gluten challenge. United European Gastroenterol J 2014; 2: 268–278.
- Vader W, Stepniak D, Bunnik EM, Kooy Y, de Haan W, Drijfhout JW, van Veelen PA, Koning F: Characterization of cereal toxicity for celiac disease patients based on protein homology in grains. Gastroenterology 2003; 125: 1105–1113.
- van de Wal Y, Kooy Y, van Veelen P, Pena S, Mearin L, Papadopulos G, Koning F: Cutting edge: selective deamidation by tissue transglutaminase strongly enhances gliadin-specific T cell reactivity. J Immunol 1998; 161: 1585–1588.
- Molberg O, Uhlen AK, Jensen T, Flaete NS, Fleckenstein B, Arentz-Hansen H, Raki M, Lundin KE, Sollid LM: Mapping of gluten Tcell epitopes in the bread wheat ancestors: implications for celiac disease. Gastroenterology 2005; 128: 393–401.
- Mitea C, Salentijn EM, van Veelen P, Goryunova SV, van der Meer IM, van den Broeck HC, Mujico JR, Monserrat V, Gilissen LJ, Drijfhout JW, Dekking L, Koning F, Smulders MJ: A universal approach to eliminate antigenic properties of alpha-gliadin peptides in celiac disease. PLoS One 2010; 5: e15637.
- Qiao SW, Ráki M, Gunnarsen KS, Løset GÅ, Lundin KE, Sandlie I, Sollid LM: Posttranslational modification of gluten shapes TCR usage in celiac disease. J Immunol 2011; 187: 3064–3071.
- Qiao SW, Christophersen A, Lundin KE, Sollid LM: Biased usage and preferred pairing of α- and β-chains of TCRs specific for an immunodominant gluten epitope in coeliac disease. Int Immunol 2014; 26: 13–19.
- Broughton SE, Petersen J, Theodossis A, Scally SW, Loh KL, Thompson A, van Bergen J, Kooy-Winkelaar Y, Henderson KN, Beddoe T, Tye-Din JA, Mannering SI, Purcell AW, Mc-Cluskey J, Anderson RP, Koning F, Reid HH, Rossjohn J: Biased T cell receptor usage directed against human leukocyte antigen DQ8-restricted gliadin peptides is associated with celiac disease. Immunity 2012; 37: 611–621.
- Petersen J, Montserrat V, Mujico JR, Loh KL, Beringer DX, van Lummel M, Thompson A, Mearin ML, Schweizer J, Kooy-Winkelaar Y, van Bergen J, Drijfhout JW, Kan WT, La Gruta NL, Anderson RP, Reid HH, Koning F, Rossjohn J: T-cell receptor recognition of HLA-DQ2-gliadin complexes associated with celiac disease. Nat Struct Mol Biol 2014; 21: 480–488.
- Petersen J, van Bergen J, Loh K, Kooy-Winkelaar Y, Beringer DX, Thompson A, Bakker SF, Mulder CJ, Ladell K, McLaren JE, Price DA, Rossjohn J, Reid HH, Koning F: Determinants of gliadin-specific T cell selection in celiac disease. J Immunol 2015; 194: 6112–6122.
- Arentz-Hansen H, Fleckenstein B, Molberg Ø, Scott H, Koning F, Jung G, Roepstorff P, Lundin KE, Sollid LM: The molecular basis for oat intolerance in patients with celiac disease. PLoS Med 2004; 1:e1.
- Janatuinen EK, Pikkarainen PH, Kemppainen TA, Kosma VM, Järvinen RM, Uusitupa MI, Julkunen RJ: A comparison of diets with and without oats in adults with celiac disease. N Engl J Med 1995; 33: 1033–1037.
- Maiuri L, Ciacci C, Ricciardelli I, Vacca L, Raia V, Auricchio S, Picard J, Osman M, Quaratino S, Londei M: Association between innate response to gliadin and activation of pathogenic T cells in coeliac disease. Lancet 2003; 362: 30–37.
- Barone MV, Troncone R, Auricchio S: Gliadin peptides as triggers of the proliferative and stress/innate immune response of the celiac small intestinal mucosa. Int J Mol Sci 2014; 15: 20518–20537.
- Koning F: Celiac disease: across the threshold of tolerance; in Fasano A, Troncone R, Brandski D (eds): Frontiers in Celiac Disease. Pediatric Adolesc Med. Basel, Karger, 2008, vol 12, pp 82–88.
- ten Dam M, Van De Wal Y, Mearin ML, Kooy Y, Pena S, Drijfhout JW, Koning F, Van Tol M: Anti-alpha-gliadin antibodies (AGA) in the serum of coeliac children and controls recognize an identical collection of linear epitopes of alpha-gliadin. Clin Exp Immunol 1998; 114: 189–195.
- van Herpen TWJM, Goryunova SV, van der Schoot J, Mitreva M, Salentijn E, Vorst O, Schenk M, van Veelen PA, Koning F, van Soest L, Vosman B, Bosch D, Hamer RJ, Gilissen LJWJ, Smulders MJM: Alpha-gliadin genes from the A, B, and D genomes of wheat contain different sets of celiac disease epitopes. BMC Genomics 2006; 7: 1.
- Spaenij-Dekking L, Kooy-Winkelaar Y, van Veelen P, Drijfhout JW, Jonker H, van Soest L, Smulders MJM, Bosch D, Gilissen LJWJ, Koning F: Natural variation in toxicity of wheat accessions for celiac disease patients. Potential for selection and breeding of nontoxic wheat varieties. Gastroenterology 2005; 129: 797–806.
- Stepniak D, Spaenij-Dekking L, Mitea C, Moester M, de Ru A, Baak-Pablo R, van Veelen P, Edens L, Koning F: Highly efficient gluten degradation with a newly identified prolyl endoprotease: implications for celiac disease. Am J Physiol Gastrointest Liver Physiol 2006; 291:G621–G629.
- Shan L, Marti T, Sollid LM, Gray GM, Khosla C: Comparative biochemical analysis of three bacterial prolyl endopeptidases: implications for coeliac sprue. Biochem J 2004; 383: 311–318.
- Gass J, Khosla C: Prolyl endopeptidases. Cell Mol Life Sci 2007; 64: 345–355.
- Mitea C, Havenaar R, Drijfhout JW, Edens L, Dekking L, Koning F: Efficient degradation of gluten by a prolyl endoprotease in a gastrointestinal model: implications for celiac disease. Gut 2008; 57: 25–32.
- Tack GJ, van de Water JM, Bruins MJ, Kooy-Winkelaar EM, van Bergen J, Bonnet P, Vreugdenhil AC, Korponay-Szabo I, Edens L, von Blomberg BM, Schreurs MW, Mulder CJ, Koning F: Consumption of gluten with gluten-degrading enzyme by celiac patients: a pilot-study. World J Gastroenterol 2013; 19: 5837–5847.
- Salden BN, Monserrat V, Troost FJ, Bruins MJ, Edens L, Bartholomé R, Haenen GR, Winkens B, Koning F, Masclee AA: Randomised clinical study: Aspergillus niger-derived enzyme digests gluten in the stomach of healthy volunteers. Aliment Pharmacol Ther 2015; 42: 273–285.
- Gass J, Bethune MT, Siegel M, Spencer A, Khosla C: Combination enzyme therapy for gastric digestion of dietary gluten in patients with celiac sprue. Gastroenterology 2007; 133: 472–480.
- Montserrat V, Bruins MJ, Edens L, Koning F: Influence of dietary components on Aspergillus niger prolyl endoprotease mediated gluten degradation. Food Chem 2015; 174: 440–445.